Research Areas
I have had a long-standing interest in how peripheral membrane enzymes bind to membranes and search out their specific targets, and how these events are modulated by cellular conditions. Very specific and tight interactions with phospholipids can occur via specific domains that anchor proteins to cell membranes. However, many proteins bind to membranes without such domains. In such cases, the canonical view is that cationic residues on the protein direct it to negatively charged membranes where hydrophobic side-chains then insert into the bilayer. Our recent work with secreted bacterial virulence factors, the phosphatidylinositol-specific phospholipase C enzymes, elaborated and modified this picture by identifying a new binding mode, phosphatidylcholine-cation – protein-π interactions, that is likely to be prominent for many amphitrophic proteins. Aside from mutagenesis, bulk phospholipid binding studies (usually by FCS and in collaboration with Anne Gershenson at U. Mass. Amherst), and kinetics of these enzymes, a major emphasis in my laboratory was to identify and determine the location of specific non-substrate phospholipid sites on proteins.
For this we used high resolution field cycling 31P and 13C relaxometry, a technique we call PR1E (paramagnetic relaxation enhancement of the R1) developed in collaboration with Dr. Alfred Redfield at Brandeis University. At very low fields (< 0.02 T), the spin-labeled protein relaxes any nearby phospholipid 31P (and potentially 13C) that stay anchored on the protein for at least the correlation time of the overall lipid aggregate. Since only the relaxation is done at low fields, we can define site specificity using a mixture of different phospholipids. Thus far, high resolution relaxometry on ‘Al’s Spin Spa’ has provided constraints for specific lipid binding sites on the PLCδ1 EF-hand domain, the Akt1 PH domain and PTEN that could be coupled with modeling to identify discrete phospholipid binding sites. |
|
A different area of research has been in identifying the array of small organic molecules that extremophiles (and even mammalian cells) synthesize in response to different stresses. Using
13C and
15N pulse NMR experiments we catalogued the variety of solutes that many archaea accumulate in response to thermal or salt stress and elucidated biosynthetic pathways. Unusual solutes include ß-amino acids (ß-glutamate, ß-glutamine,
NΕ-acetyl-ß-lysine) and carbohydrates such as sulfotrehalose and di-myo-inositol-1,1’-phosphate. With the same type of approaches applied to mammalian cell lines we identified itaconic acid (interestingly, a synthon in the polymer world) as a solute synthesized in large quantities in macrophages and macrophage-derived cell lines upon activation. It is up to others to determine exactly what all that itaconate does!
First use of high resolution relaxometry for 31 P- PR1E measurements:
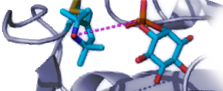
The first system explored was a bacterial PI-PLC (from Bacillus thuringiensis). We looked at the effect of different spin-labels on the protein on 31P relaxation of phosphatidylcholine (PC) and phosphatidylmethanol (PMe) in the same vesicle. Selected data below show there are discrete sites (lasting at least a µs) for each phospholipid. PMe binds in the active site and is near W47 and not as close to D205. (The spin-label on N168C is a control and shows similar behavior to these vesicles with unlabeled protein.) PC is quite close to D205 and far from W47. The rP-e values extracted from the field cycling data suggested a site on the periphery of the protein that had many Tyr residues . . . a region where PC cation – Tyr π complexes could form.
Evidence to support a discrete binding pocket for PC was supplied by MD simulations (Nathalie Reuter and her group in Bergen, Norway). Over a 300 ns time-course many transient PC-Tyr interactions were observed, particularly in helix G. Only one such PC site in the helix G region, involving Tyr246, was persistent. This was consistent with the probable location of a tight PC site as determined by field cycling 31P-NMR.
.jpg)
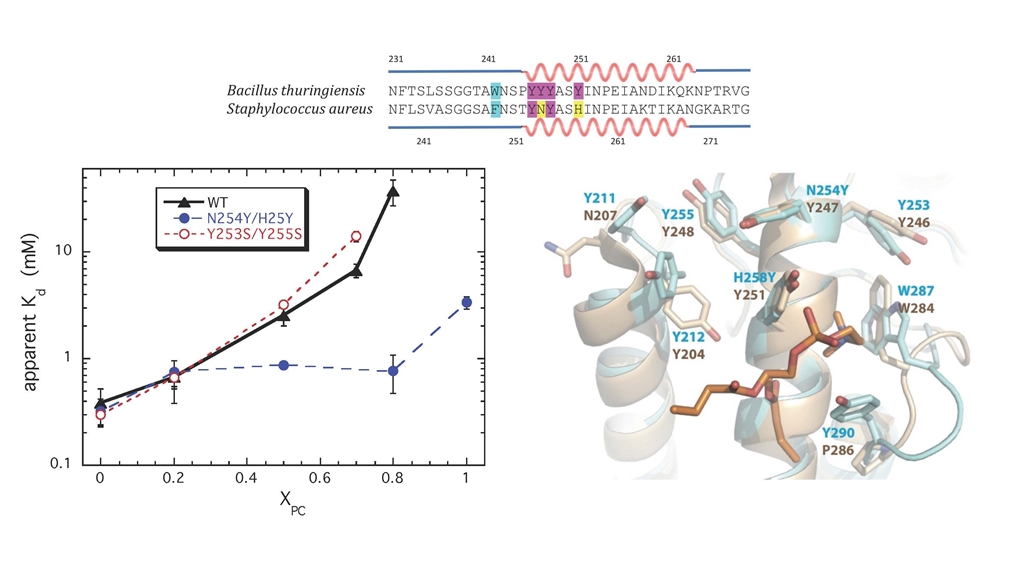
As a test of the contribution of a PC-cation – protein π interaction to membrane binding, we turned to the PI-PLC from Staphylococcal aureus. That protein has very poor binding to PC bilayers and interestingly does not have two of the four Tyr in helix G that are critical for binding in B. thuringiensis PI-PLC. Adding two more generated a protein, S. aureus N254Y/H258Y that could now bind to PC -rich membranes. We also obtained a crystal structure of S. aureus PI-PLC N254Y/H258Y (blue) with dibutyroyl-PC (orange) bound. This structure is superimposed on the B. thuringiensis PI-PLC (tan) structure. Note the plethora of aromatic rings that could engage in transient cation-π interactions with the membrane.
Use of high resolution 31P-PR1E to define phospholipid binding sites on other peripheral membrane proteins:
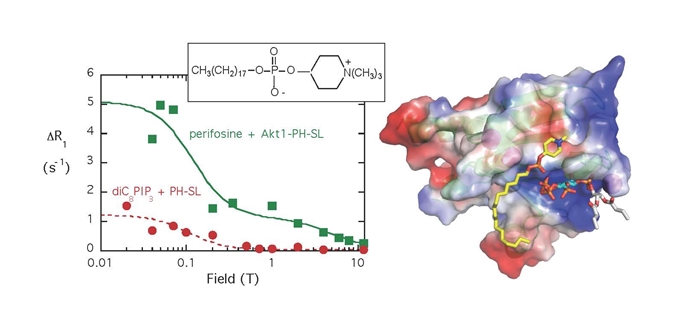
- The Akt PH domain binds cytotoxic alkyphospholipids (e. g., perifosine) in a site adjacent to but not in the PI(3, 4, 5)P3 that attaches Akt to the membrane for phosphorylation (molecule in model has the white acyl chains). The PR1E is shown for both perifosine and diC8PI(3,4,5)P3 binding to the spin-labeled PH domain (here the R1 for the lipids in the presence of unlabeled protein is subtracted). Using the NMR rp-e with molecular modeling allowed us to locate the lipid head groups on the protein. If perifosine (shown with the yellow alkyl tail) binds, it will misalign the protein on membranes and prevent tight binding and subsequent phosphorylation.
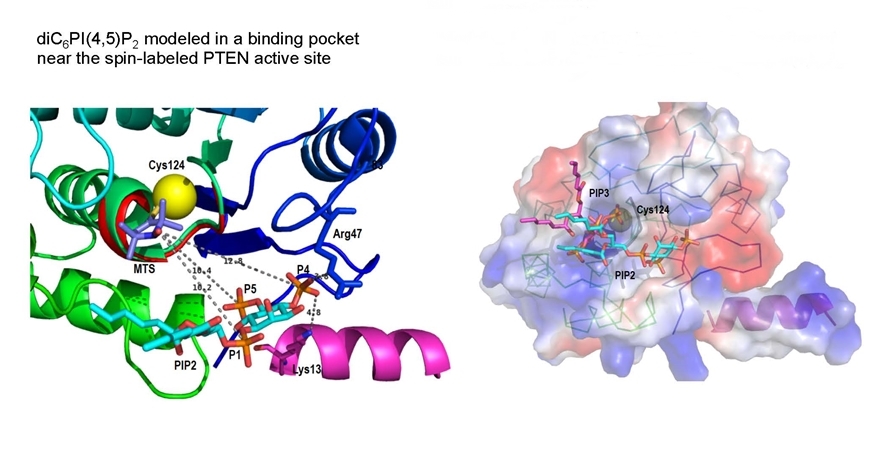
- PTEN, a tumor suppressor whose role is to hydrolyze the PI(3,4,5)P3 needed for cell growth, has 10 Cys, most of whom are spin-labeled. However the r–6 dependence of the paramagnetic relaxation means only close labels will relax the phospholipid 31P nuclei of bound molecules. Analysis of R1 versus magnetic field profiles for PI (which competes with the substrate) and the product PI(4,5)P2, (which ‘activates’ the enzyme) show they bind to different sites on PTEN.
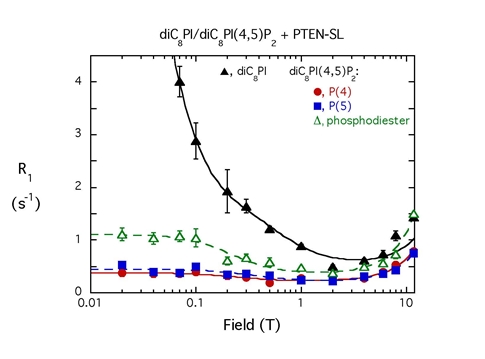 |
With micelles containing both diC8PI and diC8PI(4,5)P2, the spin-labeled PTEN is much more effective at relaxing diC8PI (which binds in the active site). The r-6 dependence means the diC8PI(4,5)P2 can’t be that far away from the active site. |
Replacing Cys124 (in the active site) with Ser in PTEN, spin-labeling the mutant, and measuring PR1E as a function of magnetic field, allowed us to use the ‘lost’ PR1E to obtain distances from the spin-label on Cys124 to bound 31P nuclei of PI and diC8PI(4,5)P2. Judicious modeling (done by Dr. Bog Stec) and theorizing that the flexible N-terminal region forms a helix (pink) when the protein binds to membranes, suggest that the product activator PI(4,5)P2 binds near the active site. The surface representation suggests how activator likely aids in processive catalysis on the membrane.
Identifying stress solutes in microorganisms:
Low molecular weight organic compounds (osmolytes or ‘compatible solutes’) are accumulated in organisms that enable cells to minimize water loss and maintain turgor pressure. Many of these solutes can also stabilize proteins to thermal denaturation. Some of the key ones we have identified in methanogens are shown below.
DIP is one of the very unusual solutes and is synthesized by many other extremophilic archaea (including Thermotoga maritima). In collaboration with the Osterman group at the Burnham Institute in La Jolla, the DIP biosynthetic pathway was determined and verified biochemically. Monitoring transcription of mRNA for the four enzymes in the pathway showed that the first enzyme, 1-L-myo-insoitol phosphate synthase (IPS), is the point of regulation. Transcription is dramatically upregulated by both heat and salt shock in T. maritima.